-
在全球范围内,侵袭性真菌感染的公共卫生问题日益严峻。白念珠菌(Candida albicans)是院内真菌感染的主要病原体之一,在侵袭性念珠菌病中占比超过50%[1]。2022年10月,世界卫生组织(WHO)将白念珠菌列为威胁健康的最高等级的关键优先级病原体之一[2]。同时,耐药性尤其是唑类耐药问题已成为全球重大挑战,新型抗真菌药物及新靶点的研究显得尤为重要[3-5]。
白念珠菌的甾醇生物合成通路通过合成麦角甾醇(ergosterol),维持细胞膜完整性及调节膜流动性[6,7]。该通路中ERG3基因编码的甾醇C-5脱氢酶(C-5 desaturase)催化甾醇前体转化为麦角甾醇合成的关键中间体,最终合成麦角甾醇[8],其功能缺陷可引发甾醇代谢重编程或改变药物敏感性。本研究基于CRISPR/Cas9技术构建白念珠菌ERG3基因缺失菌,通过甾醇代谢、药物敏感性、毒力和致病力等分析,旨在揭示ERG3基因缺失引发的甾醇代谢重塑规律,阐明其对唑类药物、多烯类药物敏感性、菌丝分化及致病力的调控作用,为克服真菌耐药提供潜在分子靶标。
-
气相色谱质谱联用仪(岛津GCMS-QP2020 NX),TS-100摇床(海门市其林贝尔仪器制造公司),TECAN酶标分析仪(Thermo Scientific),SW-CJ-1FD洁净工作台(苏净安泰空气技术有限公司),96 Well Thermal Cycler PCR仪(美国Applied Biosystems公司),EVOS相差倒置显微镜(AMG公司)。
-
白念珠菌标准菌株SC5314、质粒pV1093由Aaron P. Mitchell教授赠予;酵母浸膏、蛋白胨、胎牛血清(美国Gibco),酵母氮源基础(BBI),胆固醇(Diamond);D-葡萄糖、酵母基因组DNA快速抽提试剂盒、50×TAE缓冲液、核糖核酸酶A溶液购自生工生物,石油醚(30~60 ℃)、环己烷、甲醇、三氯甲烷购自国药集团。
-
ICR小鼠(雌性,6~8周龄)购自雷根生物技术有限公司(实验动物生产许可证编号:SCXK浙2024-0004)。所有小鼠置于光照和黑暗12 h交替循环室,室内环境为18~25 ℃,相对湿度为30%~70%。在整个实验过程中小鼠可自由获取食物和水,实验随机分组。所有动物实验均按照规定进行,并均获得海军军医大学医学伦理委员会批准。
-
使用质粒pV1093扩增Cas9片段及sgRNA表达片段。使用都柏林假丝酵母(Candida dubliniensis)HIS1标记的质粒pMH01/pMH02构建缺失基因片段(两端包含目标基因开放阅读框上下游各80 bp的同源序列)。通过醋酸锂转化法将各片段导入白念珠菌SC5314(HIS1营养缺陷型)后,采用菌落PCR及基因组PCR对阳性克隆进行验证。
-
过夜活化的白念珠菌在100 ml SC液体培养基中稀释至吸光度(A600)为0.2,30 ℃恒温摇床200 r/min培养8 h。结束后离心,弃上清液,PBS洗3次后记录湿菌重量。加入2.5 ml PBS和6 ml皂化剂,80 ℃水浴皂化1 h,冷却后加入6 ml石油醚萃取3次后合并萃取液,65 ℃水浴挥干,按每克湿菌重加1 ml环己烷溶解,加入5 μl 4 mg/ml胆固醇溶液作为内标,送气相色谱-质谱联用(GC-MS)检测。
-
过夜活化白念珠菌,测量A600后在YPD液体培养基中稀释至A600为0.4。稀释1 000倍后菌液加入96孔板中,每孔100 μl。将96孔板置于酶标仪中,设置程序参数为温度30 ℃,每间隔1 h测定A600。依据测得的吸光度数据,绘制时间-生长曲线,依据测定A600计算处于对数生长期的菌细胞倍增时间(TD),计算公式为:TD=t×lg2/lg(Nt/N0),其中,Nt为t时间的菌细胞数,N0为初始菌细胞数[9]。
-
将过夜培养的白念珠菌菌液离心收集,PBS洗3次后用RPMI
1640 培养基重悬,A600调至0.4并稀释1 000倍。在无菌96孔板第1列各孔中加入100 µl RPMI1640 培养基。第2列加200 µl菌液,第3~12列各加100 µl菌液。向第2列加预定浓度药物后从第2~11列依次倍比稀释。30 ℃培养,于24 h测定各孔的A600值,并计算药物抑制80%菌株生长的最低抑菌浓度(MIC80)。实验重复3次取平均值。 -
液体菌丝诱导实验:将过夜活化的白念珠菌离心后用PBS洗3次,测定菌液A600。在RPMI 1640培养基中,调整菌液浓度A600为0.4,进一步稀释500倍,在12孔板中每孔加入1 ml稀释后的菌液,置于37 ℃的恒温培养箱中培养6 h,拍照观察。
固体菌丝诱导实验:将过夜活化的白念珠菌离心后用PBS洗3次,测定菌液A600。在RPMI 1640培养基中,调整菌液浓度A600为0.1。吸取1 μl菌液,滴加在含有10% FBS的RPMI 1640培养基平皿表面。待液滴挥干后,将平皿倒置于37 ℃恒温培养箱中培养5 d,拍照观察。
-
将过夜活化的白念珠菌离心后用PBS洗3次,重悬于RPMI
1640 培养基调至1×106个/ml。在96孔板第1列加入100 μl RPMI 1640培养基,其余各孔加入100 μl菌液,37 ℃静置培养90 min。弃培养基,PBS洗3次,每孔加入100 μl RPMI 1640培养基。37 ℃继续培养24 h。弃培养基,PBS洗3次。避光加入稀释100倍的CCK-8溶液,37 ℃避光孵育1.5 h。每孔吸取100 μl上清液至新的96孔板。用酶标仪测490 nm处A值并计算生物被膜生成率:生物被膜生存率=A490实验组/A490阳性组×100%。 -
将过夜活化的白念珠菌用PBS洗涤后重悬,浓度调至1×106个/ml。每组10只小鼠,每只小鼠尾静脉注射0.2 ml菌悬液,连续观察20 d,记录其生存状况。
-
在本研究中,所有实验数据均使用GraphPad Prism 9.3.0进行绘图,使用IBM SPSS Statistics 26.0软件对数据进行统计分析。对于两独立样本间的统计学差异分析,采用t检验方法,对于缺失菌生长曲线差异分析,采用重测方差分析(Repeated Measures ANOVA),对于小鼠生存率的统计学差异分析,采用对数秩检验(Log-Rank)方法。以P<0.05认为差异有统计学意义。
-
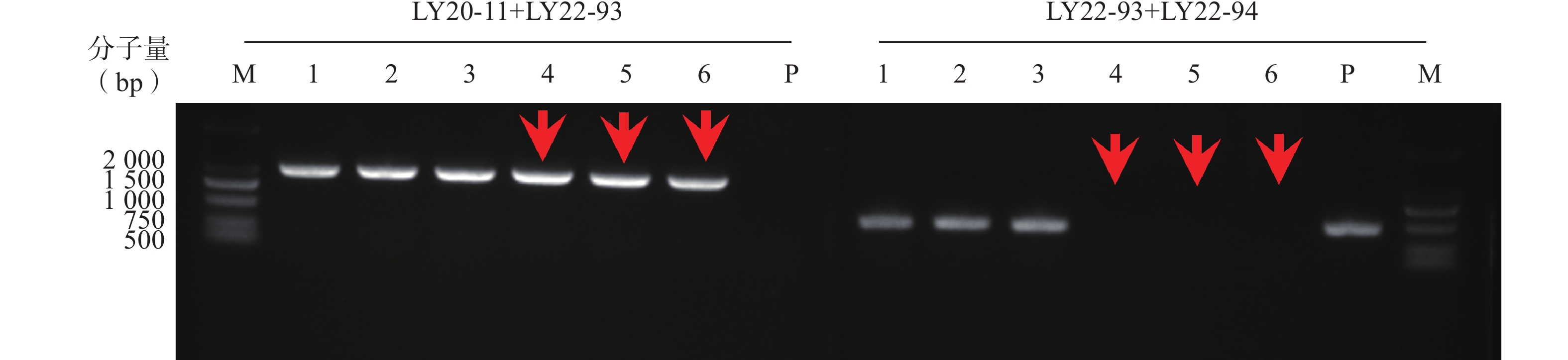
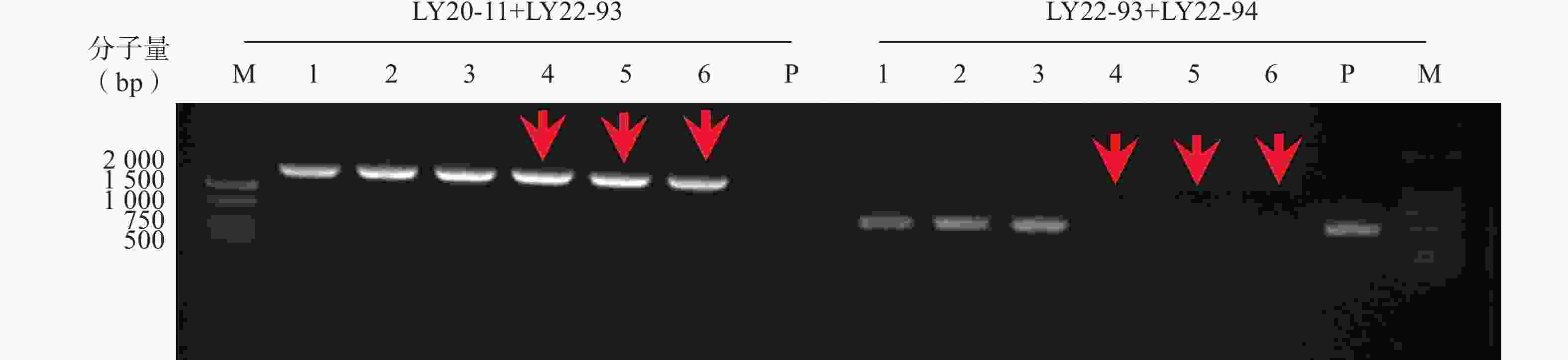
本研究通过CRISPR/Cas9技术构建ERG3基因缺失菌,通过基因组PCR验证,菌落4、5、6为ERG3双等位基因缺失菌(erg3Δ/Δ),如图1箭头所示。
-
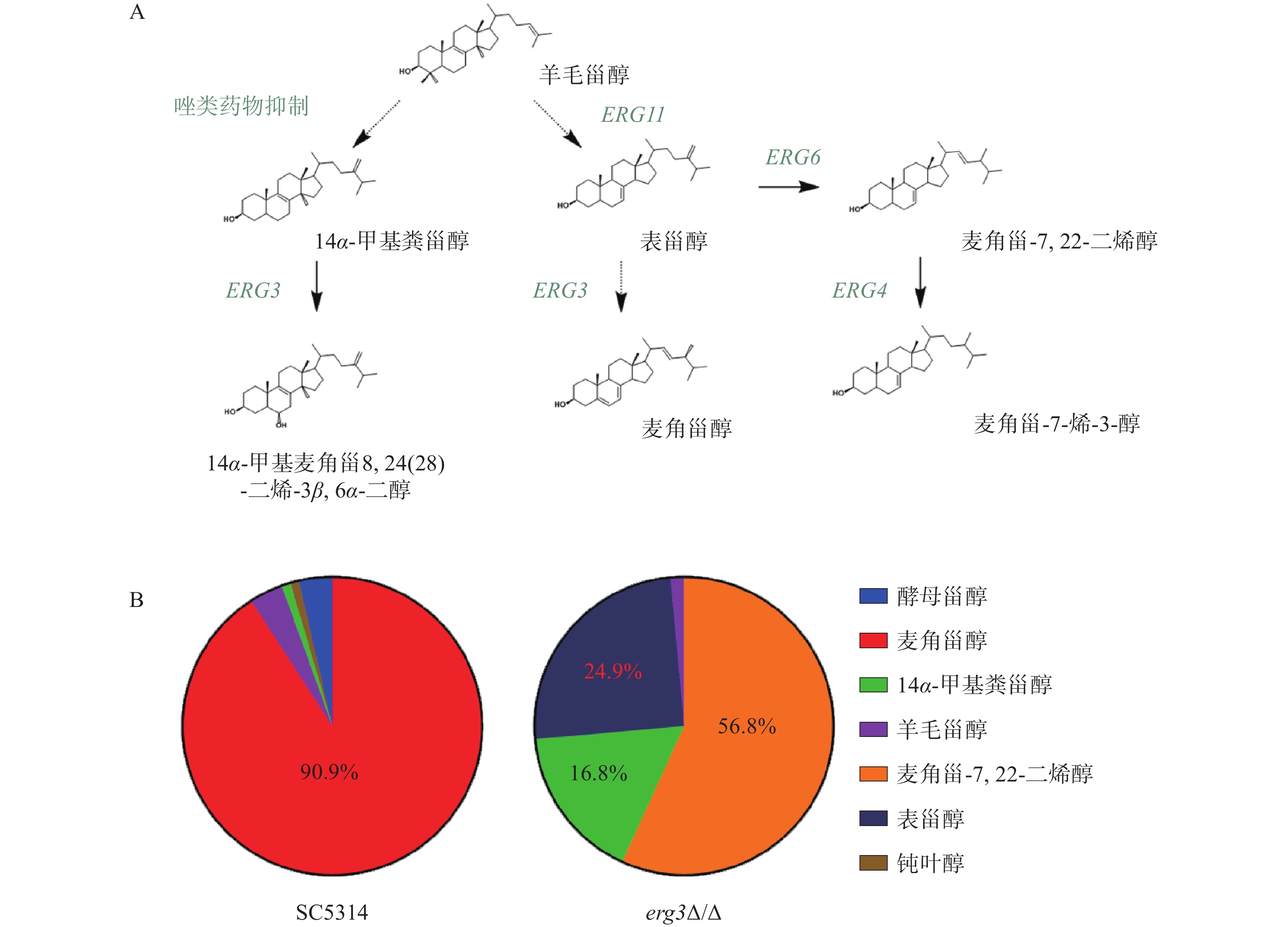
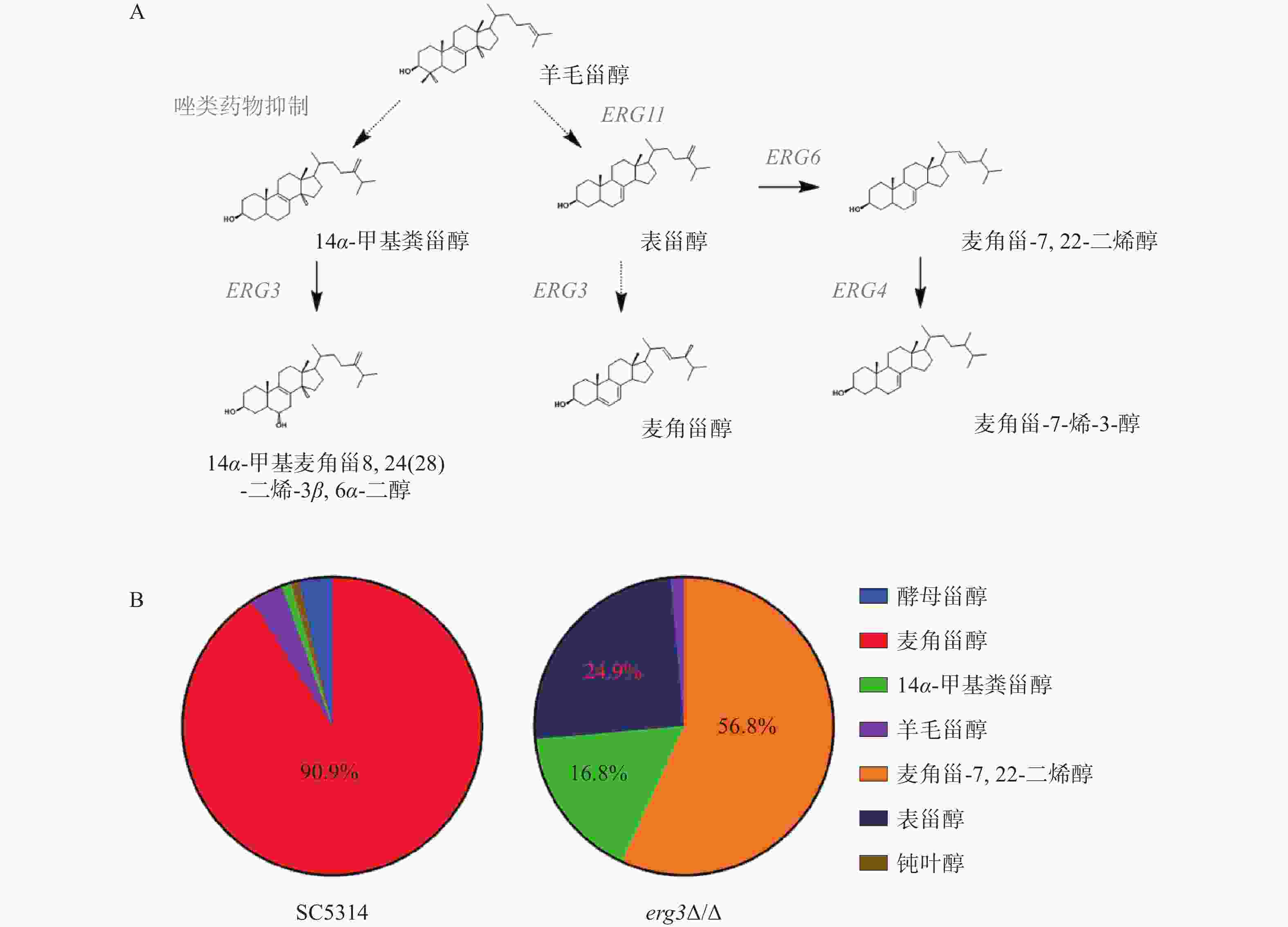
在白念珠菌甾醇生物合成通路中,ERG3基因编码的甾醇C-5脱氢酶位于该通路主路和旁路下游,在主路和旁路中其底物分别为表甾醇(episterol)和14α-甲基粪甾醇(14α-methylfecosterol),见图2A。本研究发现,亲本菌SC5314的甾醇主要成分为麦角甾醇(占比90.9%),而在ERG3缺失菌中,麦角甾-7,22-二烯醇(ergosta-7,22-dienol)、表甾醇、14α-甲基粪甾醇分别占比56.8%、24.9%和16.8%(图2B)。该结果提示,当白念珠菌ERG3基因缺失后,麦角甾醇的合成受到显著影响,ERG3编码产物C-5脱氢酶的底物表甾醇(占比24.9%)和14α-甲基粪甾醇(占比16.8%)都升高,同时,表甾醇下游因Erg3缺失无法正常产生麦角甾醇,进而从旁路Erg6催化生成大量麦角甾-7,22-二烯醇(占比56.8%),与文献报道麦角甾醇生物合成通路的下游旁路一致,进一步证明了此旁路的存在[10]。
-
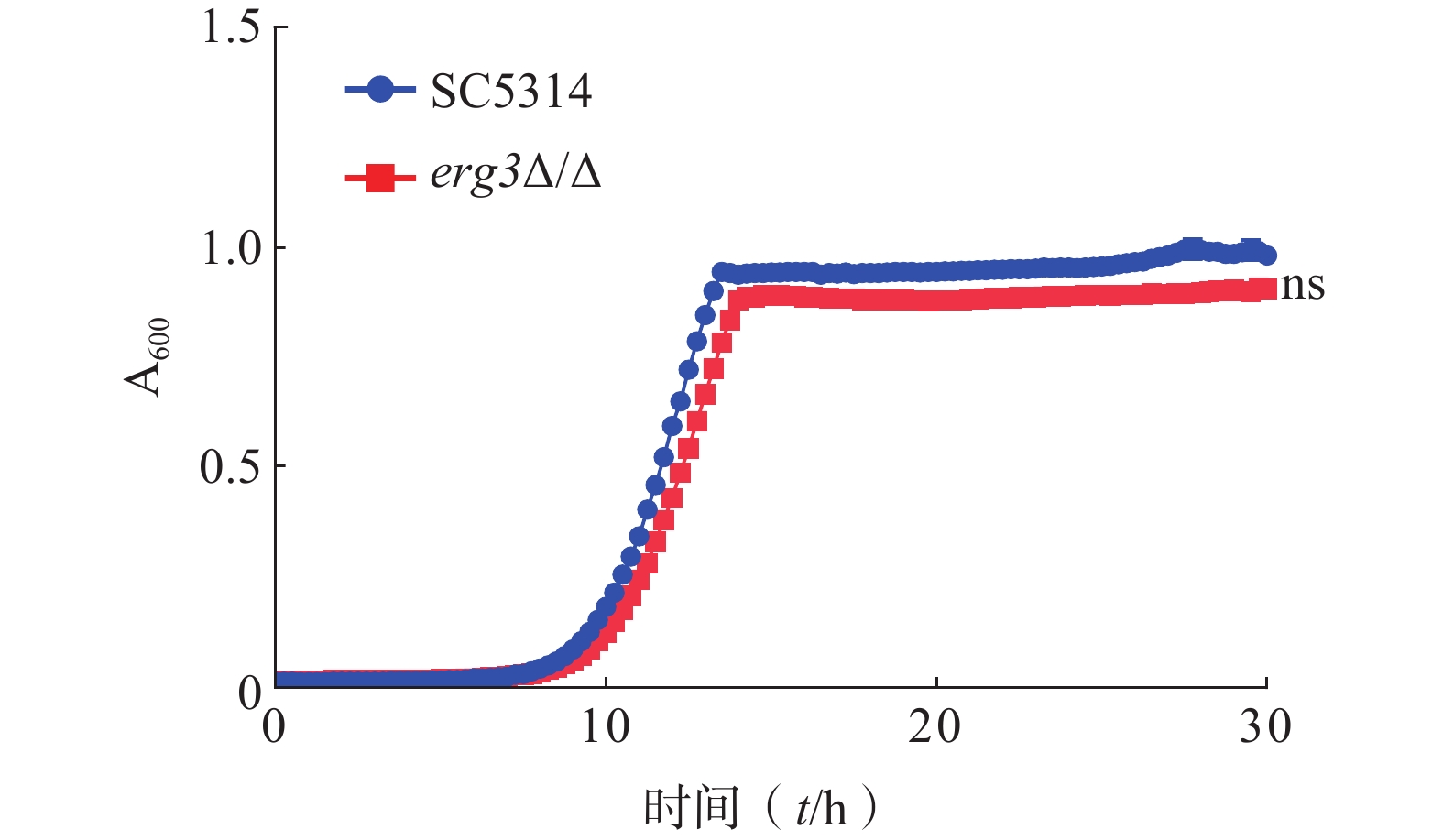
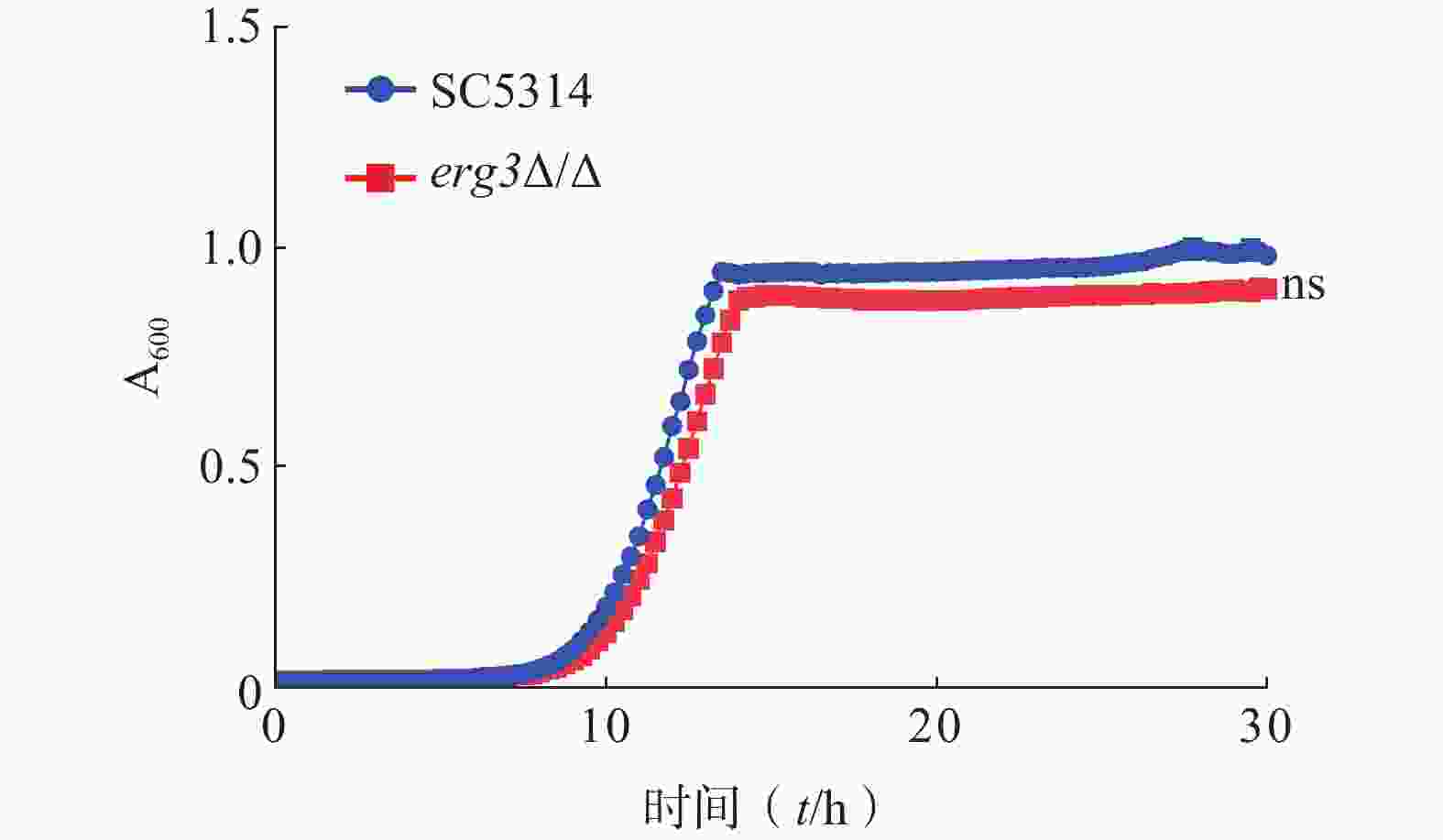
为了研究ERG3基因对白念珠菌生长的影响,本研究通过时间-生长曲线实验比较了ERG3缺失菌与SC5314的生长情况。结果显示,野生型SC5314在10~13 h达到对数生长期,倍增时间为1.13 h,ERG3缺失菌在11~14 h达到对数生长期,倍增时间为1.20 h,两者没有统计学差异(图3)。该结果提示,ERG3基因缺失对白念珠菌生长没有显著影响。
-
甾醇生物合成通路中相关基因的缺失会影响真菌对各种抗真菌药物的敏感性。本研究通过微量液基稀释法检测发现,氟康唑、伏立康唑、伊曲康唑、泊沙康唑对ERG3缺失菌的最低抑菌浓度(MIC80)均高于64 μg/ml,显著高于药物对野生型SC5314菌的MIC80值;两性霉素B对SC5314 MIC80值为1 μg/ml,对ERG3缺失菌为2 μg/ml;卡泊芬净对野生菌和ERG3缺失菌的MIC80值一致(表1)。该结果提示ERG3基因缺失可导致白念珠菌对唑类药物和多烯类药物的敏感性降低。
抗真菌药物 SC5314 erg3Δ/Δ 氟康唑 0.5 >64 伏立康唑 ≤0.125 >64 伊曲康唑 0.5 >64 咪康唑 ≤0.125 4 泊沙康唑 ≤0.125 >64 酮康唑 ≤0.125 32 两性霉素B 1 2 卡泊芬净 ≤0.031 ≤0.031 -
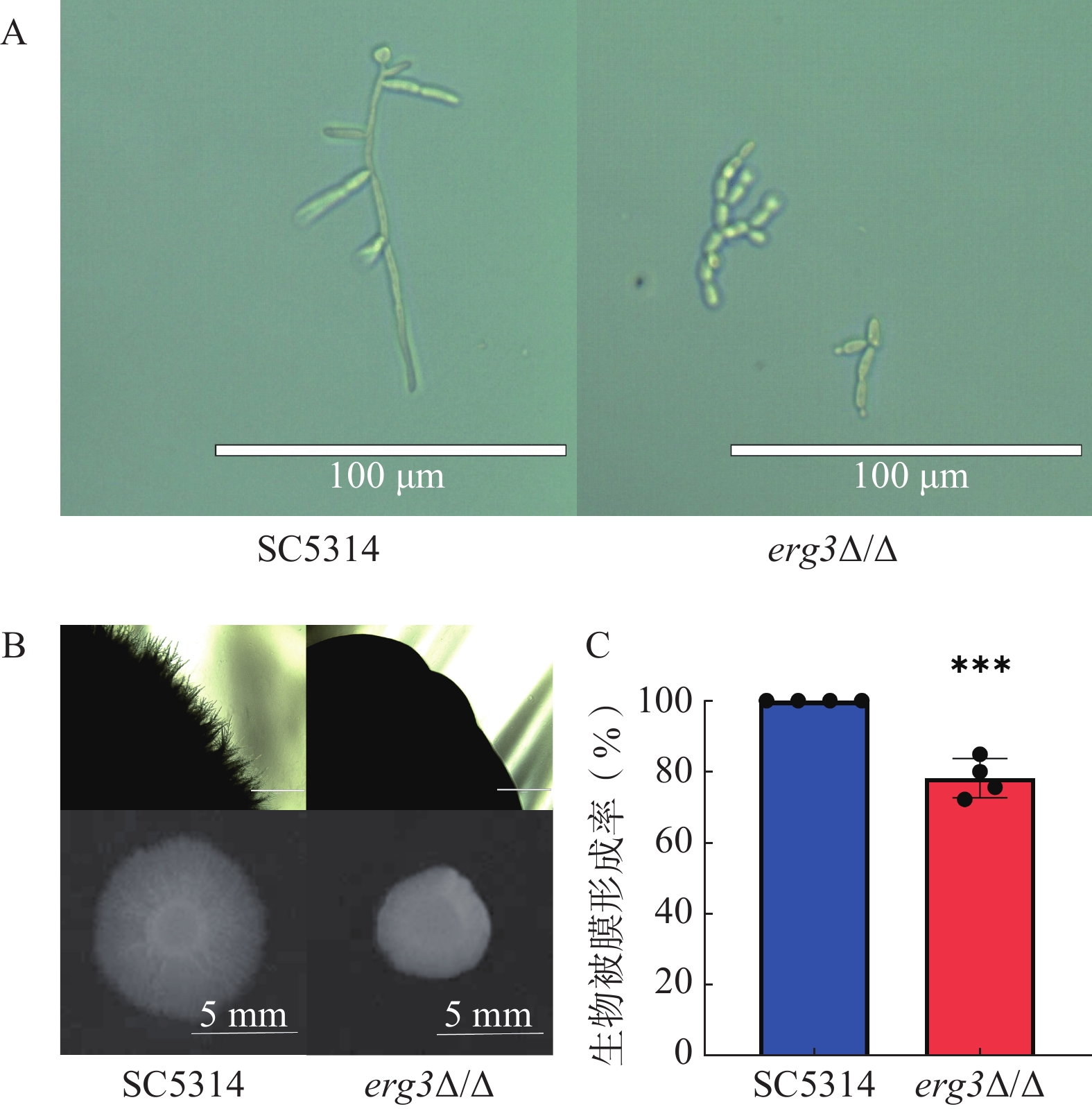
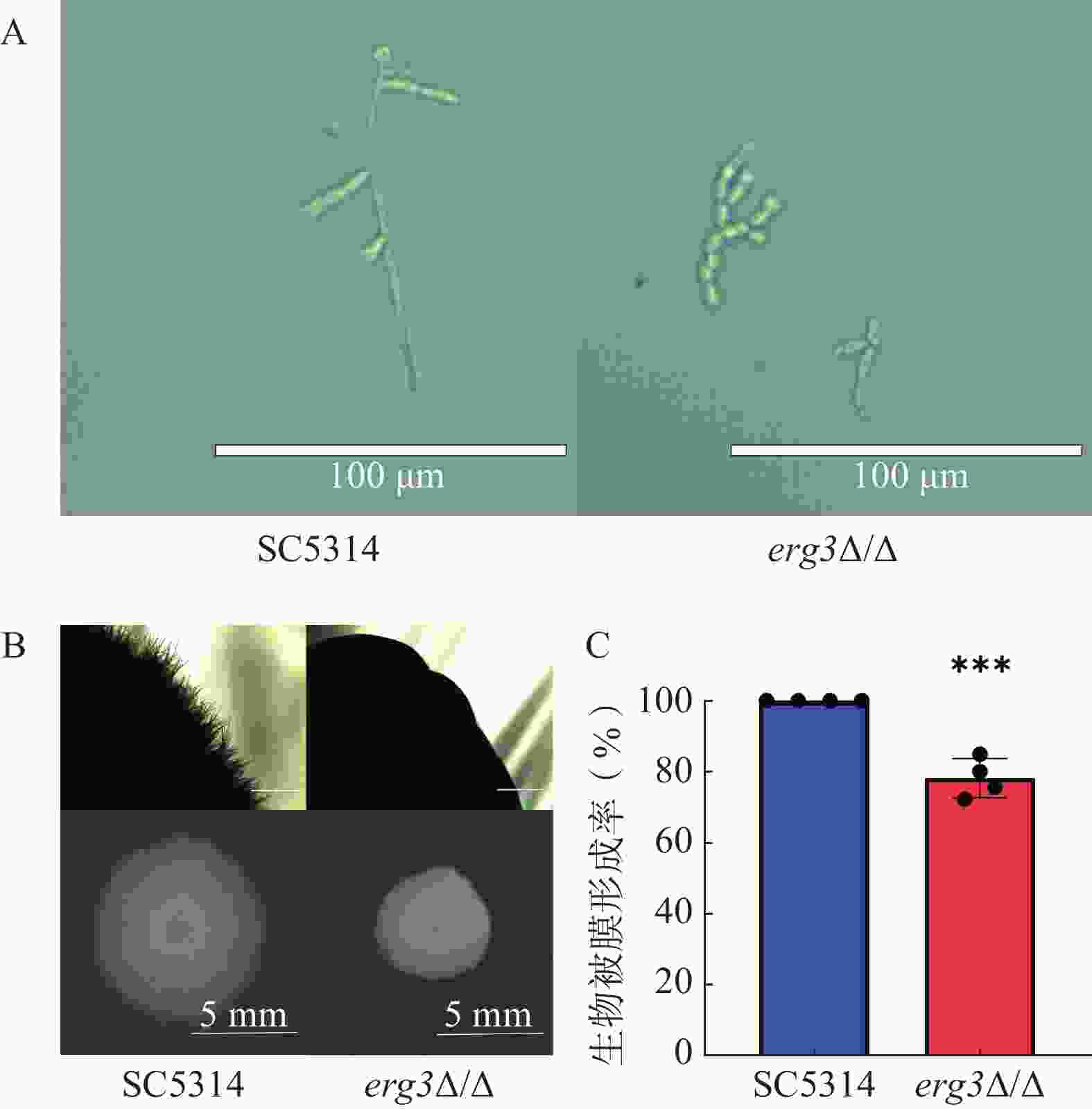
本研究探究了ERG3缺失菌在液体和固体培养基中菌丝生长情况。结果显示,在液体培养基中诱导6 h,野生型SC5314的菌丝细长且有分支,随机测量30根菌丝,其平均长度为96 μm,ERG3缺失菌的菌丝生长严重受抑,细胞几乎都呈现酵母态(图4A);在固体培养基诱导120 h,SC5314菌落直径为9.1 mm,ERG3缺失菌菌落四周无辐射状菌丝,菌落直径也显著减少为5.6 mm,这与亲本菌SC5314相比显著缺陷(图4B)。在生物被膜形成实验中,与野生型SC5314相比,ERG3缺失菌的生物被膜形成率为78.2%,表明ERG3基因缺失后白念珠菌生物被膜形成受到一定程度抑制(图4C)。以上结果表明ERG3基因缺失后,白念珠菌的菌丝生长和生物被膜形成受到了不同程度的抑制。
-
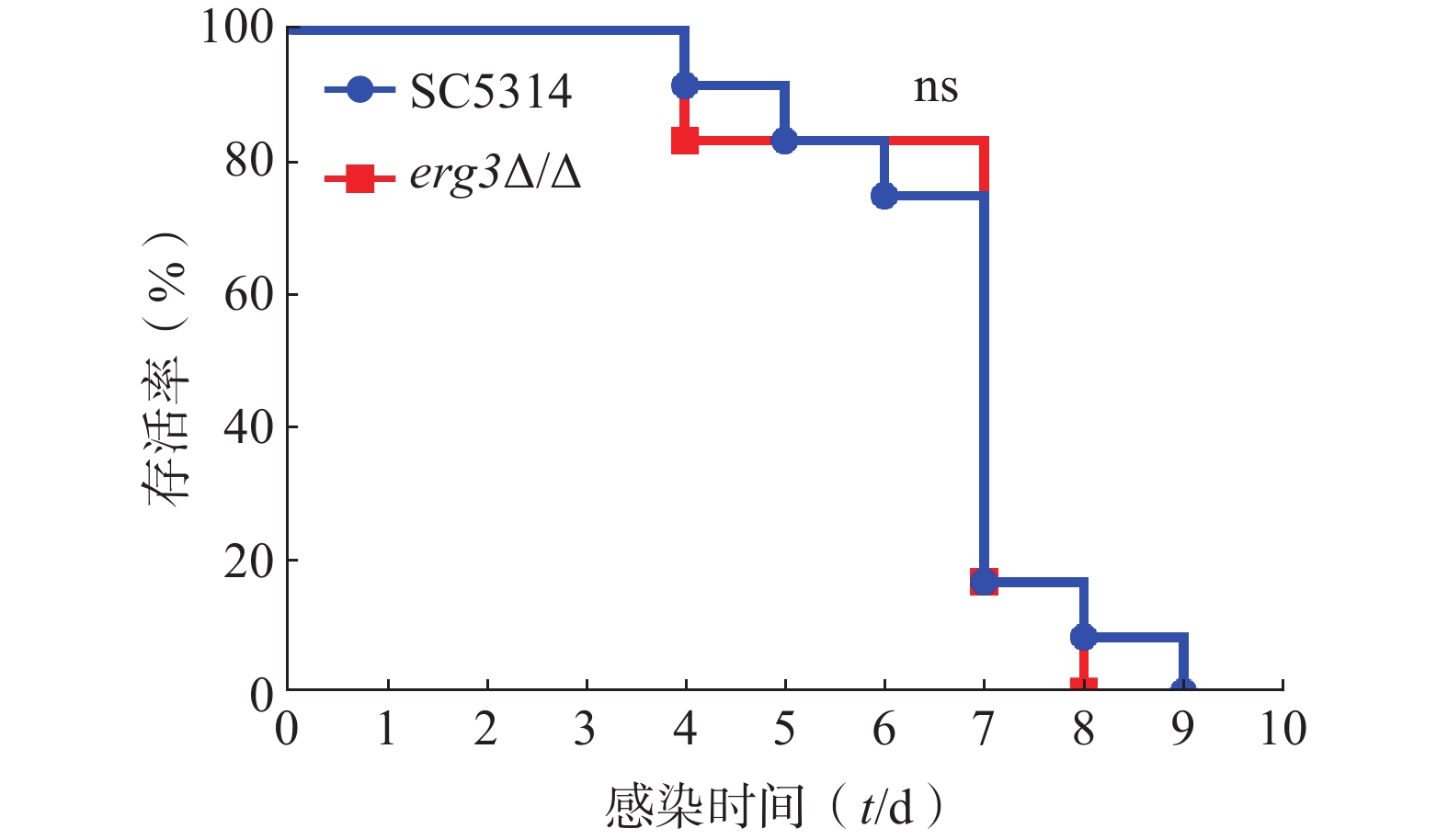
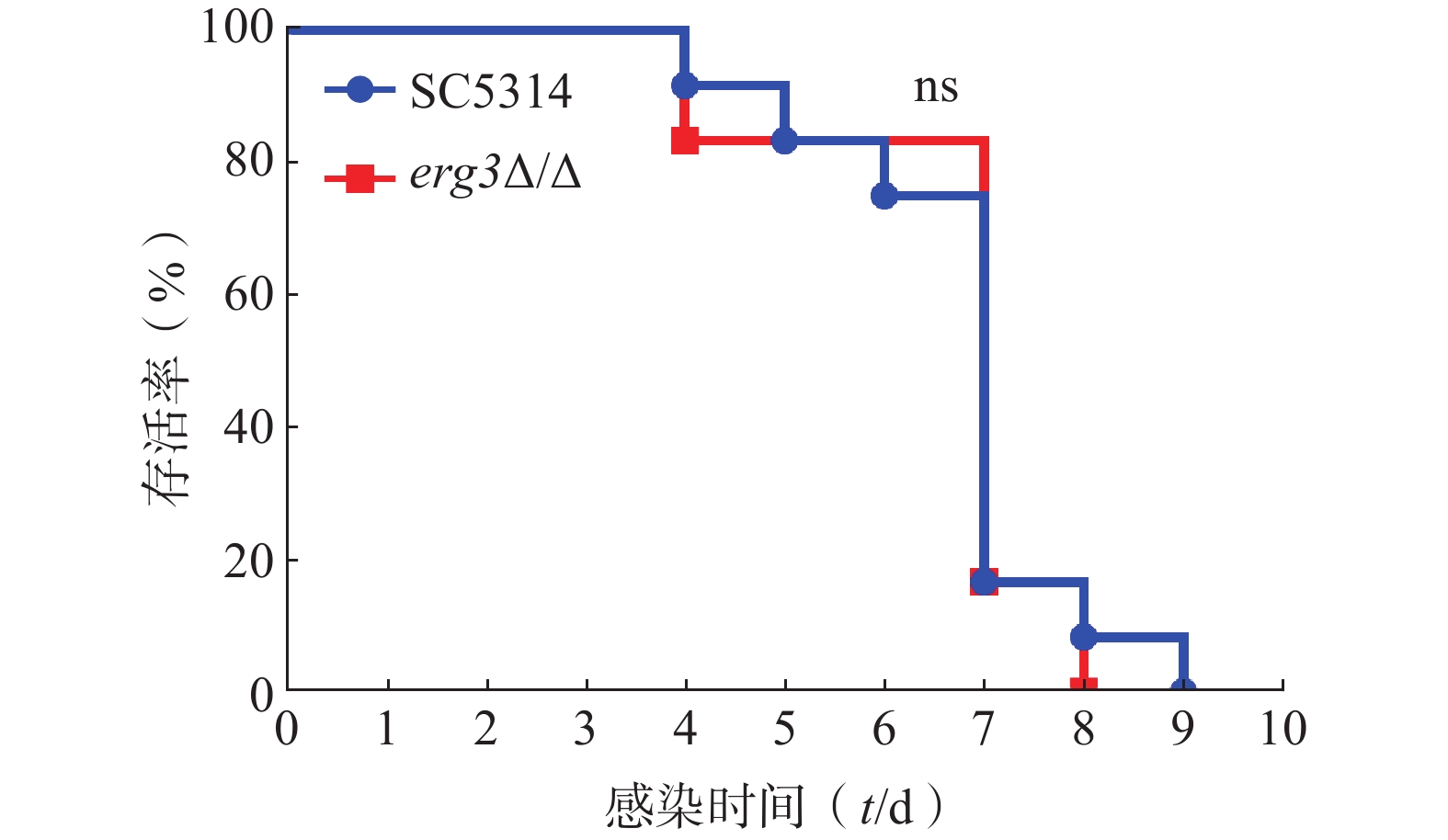
利用小鼠系统性真菌感染模型,继续探究ERG3基因缺失对白念珠菌致病力的影响。如图5所示,感染ERG3缺失菌的小鼠与感染SC5314的对照组小鼠均在第4天开始死亡,第9天全部死亡,表明ERG3基因缺失没有显著改变白念珠菌在宿主体内的致病力。
-
临床治疗侵袭性真菌感染的治疗药物主要有以下3类,通过特异性阻断真菌麦角甾醇生物合成途径的不同环节发挥作用:烯丙胺类(如特比奈芬)抑制Erg1(角鲨烯单加氧酶);多烯类(如两性霉素B)直接结合细胞膜麦角甾醇;唑类(如氟康唑)靶向抑制Erg11(C-14α-去甲基酶)。随着真菌耐药性问题日益严重,研发针对麦角甾醇合成通路其他关键酶及调控机制的新药成为抗真菌药物研究的热点。本研究通过对白念珠菌甾醇合成通路ERG3基因进行敲除,探究其对菌株甾醇代谢、生长、药物敏感性、菌丝及生物被膜形成、体内致病力的影响,旨在为抗真菌新靶标的发现奠定基础。ERG3基因的缺失并未显著影响白念珠菌的生长,推测ERG3基因缺失后,经下游旁路产生麦角甾-7,22-二烯醇,减少了甾醇中间产物的积累以维持生长,与文献报道一致[12]。ERG3基因敲除后,降低了菌株对唑类及多烯类药物的敏感性,推测因麦角甾醇合成减少导致药物靶酶含量减少从而降低敏感性。同时,ERG3基因缺失抑制菌丝及生物被膜形成,但在小鼠系统性真菌感染模型中,ERG3基因缺失对菌株致病力无显著影响,有文献报道其在小鼠肠道感染模型中为毒力必需基因,推测在小鼠肠道感染模型中,ERG3缺失菌菌丝形成能力减弱,导致菌株在肠道内的定植能力、对肠道黏膜的侵袭能力以及引发炎症反应的能力下降[13]。本研究发现ERG3基因缺失导致体外毒力减弱而体内致病力不变,潜在机制可能是:①ERG3缺失导致表甾醇、14α-甲基粪甾醇等异常甾醇富集,虽改变膜流动性,但可能通过膜脂质重构(如鞘脂类比例升高)维持膜功能;②宿主体内环境下,ERG3缺失菌可能通过膜重塑介导免疫逃逸;③ERG3缺失诱导抗氧化酶表达,抵抗宿主巨噬细胞产生活性氧的攻击等等。
综上所述,本研究为探究白念珠菌ERG3生物学功能提供实验依据。ERG3是白念珠菌麦角甾醇合成通路的关键调控因子,其缺失通过重塑甾醇代谢谱诱导多药耐药表型,但致病性维持依赖其他代偿策略,尚需要进一步开展机制研究。本研究为靶向甾醇代谢的抗真菌药物研发及耐药防控提供了理论依据。
Study on the functions of ERG3 in Candida albicans
doi: 10.12206/j.issn.2097-2024.202505044
- Received Date: 2025-05-17
- Rev Recd Date: 2025-06-20
- Available Online: 2025-09-16
- Publish Date: 2025-09-25
-
Key words:
- Candida albicans /
- sterol biosynthesis pathway /
- C-5 desaturase
Abstract:
| Citation: | YE Zi, WANG Ruina, LIU Jiacun, YANG Shiyun, LIANG Chan, YAN Lan. Study on the functions of ERG3 in Candida albicans[J]. Journal of Pharmaceutical Practice and Service, 2025, 43(9): 431-435, 454. doi: 10.12206/j.issn.2097-2024.202505044 |






 DownLoad:
DownLoad: